Carbon metabolism of intracellular bacterial pathogens and possible links to virulence
New technologies such as high-throughput methods and 13C-isotopologue-profiling analysis are beginning to provide us with insight into the in vivo metabolism of microorganisms, especially in the host cell compartments that are colonized by intracellular bacterial pathogens. In this Review, we discuss the recent progress made in determining the major carbon sources and metabolic pathways used by model intracellular bacterial pathogens that replicate either in the cytosol or in vacuoles of infected host cells.
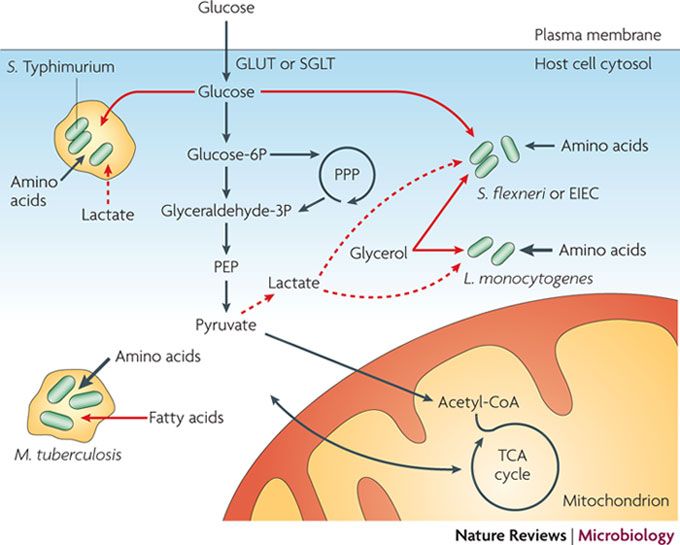
- Recent progress has expanded our knowledge about the metabolism of the model bacterial pathogens Listeria monocytogenes, Shigella flexneri (and the closely related enteroinvasive Escherichia coli (EIEC)), Salmonella enterica subsp. enterica serovar Typhimurium and Mycobacterium tuberculosiswhen living inside the host cell.
- Differences in the metabolic characteristics of these four pathogens have been elucidated in the context of the metabolism of host cell lines used for in vitro infection.
- There are several tools available to study the metabolism of these intracellular pathogens, and differential gene expression profiling (DGEP) and 13C isotopologue analysis (13C-IPA) have been particularly fruitful; however, there are both strengths and weaknesses for these techniques.
- Models have been suggested (mainly on the basis of data from DGEP and 13C-IPA studies) for the metabolic pathways and fluxes used by the four pathogens when replicating in their specific intracellular compartments (the cytosol or specific phagosomal vacuoles of the host cell). Each pathogen adapts specifically to the host cell environment but exhibits a surprisingly high metabolic flexibility in response to altered metabolic conditions.
- There is limited experimental evidence for interference by the metabolism of these intracellular bacteria with the expression of virulence genes that are required for their intracellular lifestyles.
- There is an urgent need for improved in vivo systems and more sensitive analytical tools for studying the metabolism of the bacterial pathogens in real target cells and animal models. source: https://go.nature.com/2yJmT5v
To Learn more attend The World Conference on Bacteriology and Infectious Diseases
To know more Visit: http://bit.ly/bacteriology2018
Contact us: bacteriology@pulsusevents.org
No comments:
Post a Comment